~ Project ~
Aim of this project
In order to make sure that we understand what life really is, we need to know all the required components and their functional interactions in a cell. We want to test the hypothesis that a Gram-positive bacterium can live with a defined set of just 523 and 119 protein coding and RNA genes, respectively, in a complex medium at 37°C. The implementation of this project will demonstrate that we have a good understanding of the essential processes of information processing and transfer, metabolism, cell division and cell envelope synthesis, as well as the interactions of the involved proteins. However, the project is also intended to reveal gaps in our knowledge that will be closed by detailed analyses.
The ultimate goal of the project is to design and create a Bacillus subtilis strain equipped with a minimal core genome, where all remaining genes are essential. Importantly, we want to understand for each of these genes why it has to be part of the minimal genome and how it contributes to life.
The information we expect to obtain in the course of this project can be used to develop functional genome cassettes that can be plugged together as required for the specific conditions or applications. Such de novo designed organisms would be a perfect basis for biotechnology.
Steps
Prior Work
So far, B. subtilis strains with genomes reduced by 7.7% and 20% have already been constructed. In addition, a recent genome-wide study about individually dispensable regions revealed novel unknown essential genes and functions (Westers et al., 2003; Morimoto et al., 2008; Tanaka et al., 2013). The strain deleted for all prophages and a large dispensable polyketide biosynthetic operon (genome reduction of 7.7%) serves as the starting point for this project.
I. Classification of the entire B. subtilis genome
n the basis of our functional annotation of the metabolism and our knowledge about essential genes and essential functions, each gene of B. subtilis was classified and assigned to one of the following groups: Dispensable, essential, reasonably important (because mutations cause a severe growth defect), paralogue or functionally paralogue (here a specific function, which can be performed by multiple related or non-related genes, is essential) and competence (needed for reducing the genome, not for a living cell) (Fig. 1). Thus, 642 genes, distributed in 6 functional categories, are regarded as “necessary” for MiniBacillus (Fig. 2) (Reuß et al., 2016).
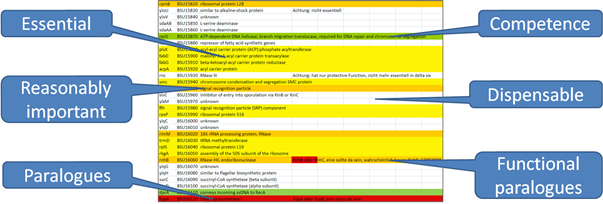
Figure 1: Exemplary classification of the B. subtilis genes
The entire B. subtilis genome has been checked; each gene was classified and assigned to one of the shown groups. Essential: A gene which cannot be deleted. Reasonably important: Mutations in these genes cause severe growth defects. Paralogue or functionally paralogue: A specific function, which can be performed by multiple related or non-related genes, is essential. Competence: This function is needed to reduce the genome, not for a living cell. Dispensable: All genes which are not assigned to one of the above mentioned groups. As a result, 610 genes are marked as necessary for the Minibacillus genome. The list is re-evaluated on a regular basis.
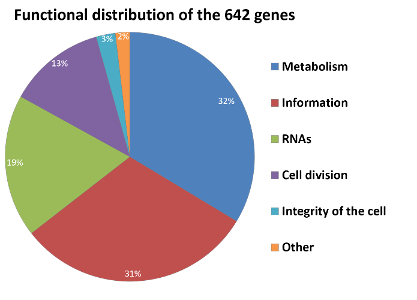
Figure 2: Functional distribution of the 642 important genes of MiniBacillus
Metabolism: Genes involved in the central metabolism, respiration, amino acid synthesis/transport, lipid synthesis, nucleotide synthesis, phosphate metabolism, synthesis of cofactors and FeS-clusters as well as acquisition of metals. Information: Genes involved in transcription, translation and posttranslational events (e.g. quality control and secretion). RNAs: rRNAs and tRNAs. Cell division: Genes required for the cell division and the cell envelope synthesis. Integrity of the cell: Genes involved in genome integrity and protection. Others: All genes with functions which are not very well understood yet.
II. Design of the minimal genome
With the classification in hand, the minimal genome will be designed by integrating knowledge from several databases and collaborations. SubtiWiki provides general information on each gene (e.g. function, location, regulatory events, operon structure, etc). Furthermore, it gives access to SubtiExpress containing detailed information on the expression of each gene based on the global analysis under different conditions, SubtiPathways with its overview of the B. subtilis metabolic network and SubtInteract visualizing all known interactions. Moreover, literature research and additional information from collaboration partners specialized in different areas of Bacillus research are integrated in specific cases (Michna et al., 2016) (Fig.3).
III. Construction of the minimal genome
The minimal genome design serves as a blueprint for the construction of the deletion plasmids (Fig. 3). A more detailed overview on the major methods used in this study can be found here. Since, we use marker-free deletions in this project we cannot transfer the genotype into other strains, therefore, only a consecutive progress is possible. Hence, a strict logistic strategy is required.
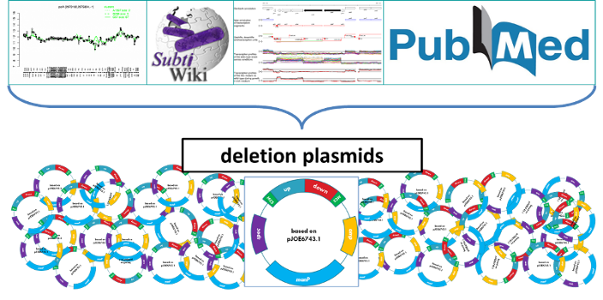
Figure 3: Integration of the comprehensive knowledge in minimal genome design and construction
The deletion of specific genomic regions requires a wide range of additional data. SubtiWiki and its several applications provides the most compact and up-to-date collection of B. subtilis research. All available information provided by this database together with literature research and input from collaborations leads to an individual design and construction of the deletion plasmids.
IV. Characterization of the minimized strains and derived suppressor mutants
The resulting minimized strains are supposed to grow in a complex medium at 37°C. Furthermore, they should still be competent and reasonably fast growing. Each strain will be characterized according to these criteria. As the project advances we expect that the strains may fail to meet these criteria. Once this happens, long-term incubations on agar plates or condition-driven selection can lead to the isolation of suppressor mutants. This process will be repeated until the strains regain the requested abilities. Genome sequencing will reveal the responsible mutations which give new insights in unknown functions, interactions and moonlighting functions.
Furthermore, we want to analyze the genome-reduced strains in multi-omics experiments (i.e. transcriptomics, proteomics, and metabolomics). These global investigations are crucial to get an insight how the strains adapt to the deletion of large genomic regions. Additionally, the results will identify promising targets for further deletions and genetic modifications.
V. Transposon mutagenesis of the final MiniBacillus
At the very end of the project, the Minibacillus will be subject to a transposon mutagenesis for a critical evaluation of the constructed genome. Obtained clones will be sequenced to identify the transposon integration in the genome. This method will reveal whether all remaining genes, which are not required for competence, recombination and DNA repair, are essential for MiniBacillus. A further genome reduction will be performed, if required, in order to obtain a strain which cannot be reduced anymore in a reasonable manner.
Essential genes
You can find a full list of identified essential genes in B. subtilis here.

